前置文章:
将一维机械振动信号构造为训练集和测试集(Python)
https://mp.weixin.qq.com/s/DTKjBo6_WAQ7bUPZEdB1TA
旋转机械振动信号特征提取(Python)
https://mp.weixin.qq.com/s/VwvzTzE-pacxqb9rs8hEVw
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from matplotlib.colors import ListedColormap
import matplotlib.patches as mpatches
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from sklearn import tree
import joblib df_train = pd.read_csv("statistics_10_train.csv" , sep = ',')
df_test = pd.read_csv("statistics_10_test.csv" , sep = ',')X_train = df_train[['Kurtosis', 'Impulse factor', 'RMS', 'Margin factor', 'Skewness','Shape factor', 'Peak to peak', 'Crest factor']].values
y_train = df_train['Tipo'].values
X_test = df_test[['Kurtosis', 'Impulse factor', 'RMS', 'Margin factor', 'Skewness','Shape factor', 'Peak to peak', 'Crest factor']].values
y_test = df_test['Tipo'].valuesmax_depth_values = range(1, 20)
scores_train = []
scores_test = []
for m in max_depth_values:treeModel = tree.DecisionTreeClassifier(random_state = 0, max_depth = m)treeModel.fit(X_train, y_train)scores_train.append(treeModel.score(X_train, y_train))scores_test.append(treeModel.score(X_test, y_test))
plt.figure()
plt.xlabel('max_depth')
plt.ylabel('Accuracy')
plt.plot(max_depth_values, scores_train, label = 'Train')
plt.plot(max_depth_values, scores_test, label = 'Test')
plt.legend()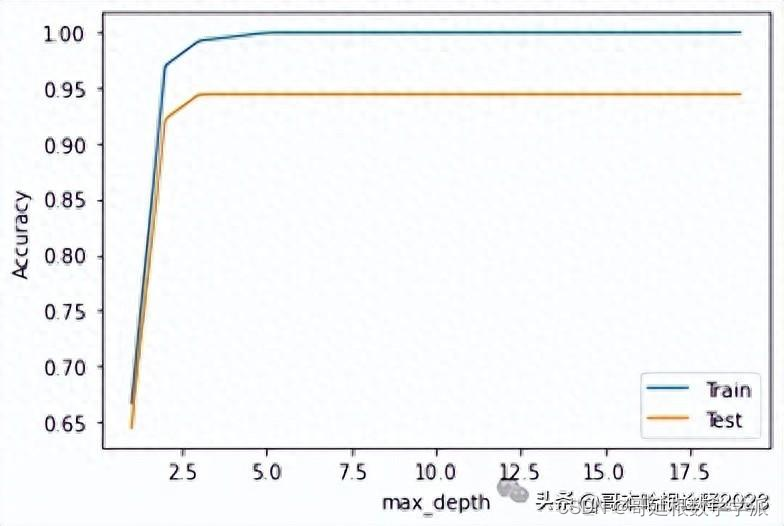
treeModel = tree.DecisionTreeClassifier(random_state = 0, max_depth = 7)
treeModel.fit(X_train, y_train)DecisionTreeClassifier(max_depth=7, random_state=0)tree.plot_tree(treeModel) [Text(200.88000000000002, 199.32, 'X[6] <= 0.14\ngini = 0.667\nsamples = 270\nvalue = [90, 90, 90]'),Text(167.40000000000003, 163.07999999999998, 'X[0] <= 3.726\ngini = 0.5\nsamples = 180\nvalue = [0, 90, 90]'),Text(66.96000000000001, 126.83999999999999, 'X[4] <= 0.397\ngini = 0.12\nsamples = 94\nvalue = [0, 6, 88]'),Text(33.480000000000004, 90.6, 'gini = 0.0\nsamples = 87\nvalue = [0, 0, 87]'),Text(100.44000000000001, 90.6, 'X[6] <= 0.055\ngini = 0.245\nsamples = 7\nvalue = [0, 6, 1]'),Text(66.96000000000001, 54.359999999999985, 'gini = 0.0\nsamples = 6\nvalue = [0, 6, 0]'),Text(133.92000000000002, 54.359999999999985, 'gini = 0.0\nsamples = 1\nvalue = [0, 0, 1]'),Text(267.84000000000003, 126.83999999999999, 'X[2] <= 3.032\ngini = 0.045\nsamples = 86\nvalue = [0, 84, 2]'),Text(234.36, 90.6, 'X[5] <= 665.031\ngini = 0.023\nsamples = 85\nvalue = [0, 84, 1]'),Text(200.88000000000002, 54.359999999999985, 'X[6] <= 0.062\ngini = 0.245\nsamples = 7\nvalue = [0, 6, 1]'),Text(167.40000000000003, 18.119999999999976, 'gini = 0.0\nsamples = 6\nvalue = [0, 6, 0]'),Text(234.36, 18.119999999999976, 'gini = 0.0\nsamples = 1\nvalue = [0, 0, 1]'),Text(267.84000000000003, 54.359999999999985, 'gini = 0.0\nsamples = 78\nvalue = [0, 78, 0]'),Text(301.32000000000005, 90.6, 'gini = 0.0\nsamples = 1\nvalue = [0, 0, 1]'),Text(234.36, 163.07999999999998, 'gini = 0.0\nsamples = 90\nvalue = [90, 0, 0]')]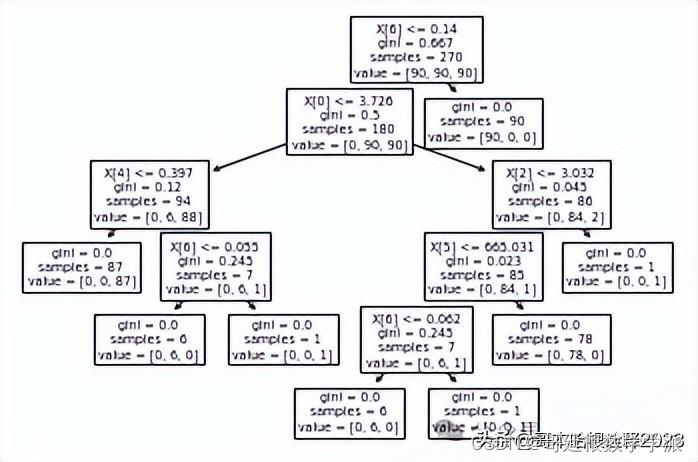
target_names = ['Inner', 'Outer', 'Healthy']
pred = treeModel.predict(X_test)
print(confusion_matrix(y_test, pred))
print(classification_report(y_test, pred, target_names = target_names))[[28 1 1][ 0 30 0][ 0 3 27]]precision recall f1-score supportInner 1.00 0.93 0.97 30Outer 0.88 1.00 0.94 30Healthy 0.96 0.90 0.93 30accuracy 0.94 90macro avg 0.95 0.94 0.94 90
weighted avg 0.95 0.94 0.94 90pred_train = treeModel.predict(X_train)
print(confusion_matrix(y_train, pred_train))
print(classification_report(y_train, pred_train, target_names = target_names))[[90 0 0][ 0 90 0][ 0 0 90]]precision recall f1-score supportInner 1.00 1.00 1.00 90Outer 1.00 1.00 1.00 90Healthy 1.00 1.00 1.00 90accuracy 1.00 270macro avg 1.00 1.00 1.00 270
weighted avg 1.00 1.00 1.00 270sns.set()
mat = confusion_matrix(y_test, pred)
fig, ax = plt.subplots(figsize=(7,6))
sns.set(font_scale=1.3)
sns.heatmap(mat.T, square=False, annot=True, fmt='d', cbar=False,xticklabels=['Fallo inner race', 'Fallo oute race', 'Healthy'],yticklabels=['Fallo inner race', 'Fallo oute race', 'Healthy'],cmap=sns.cubehelix_palette(light=1, as_cmap=True))plt.xlabel('true label');
plt.ylabel('predicted label');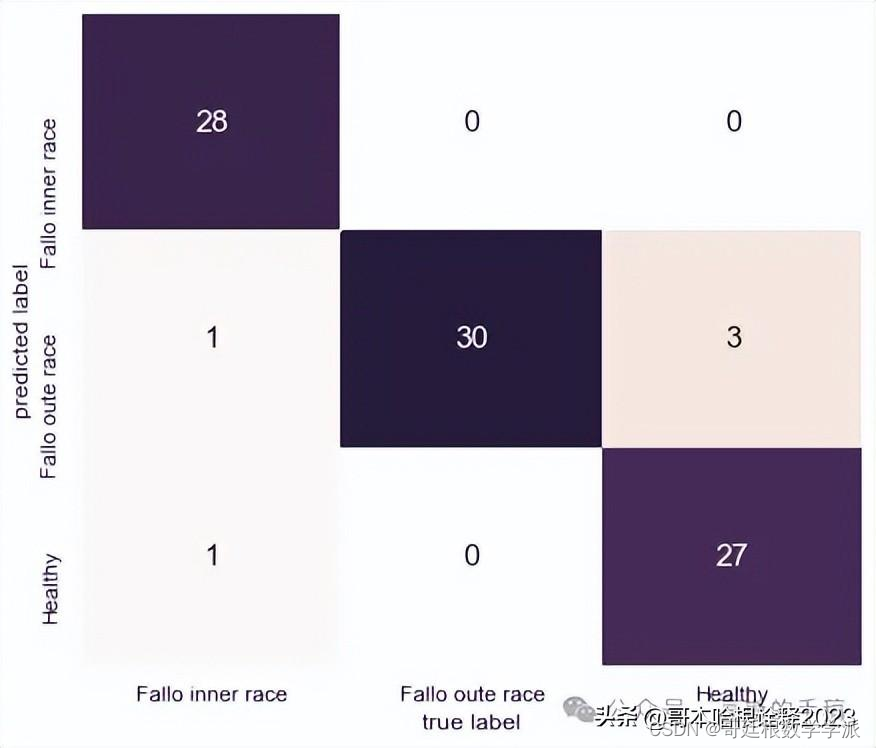
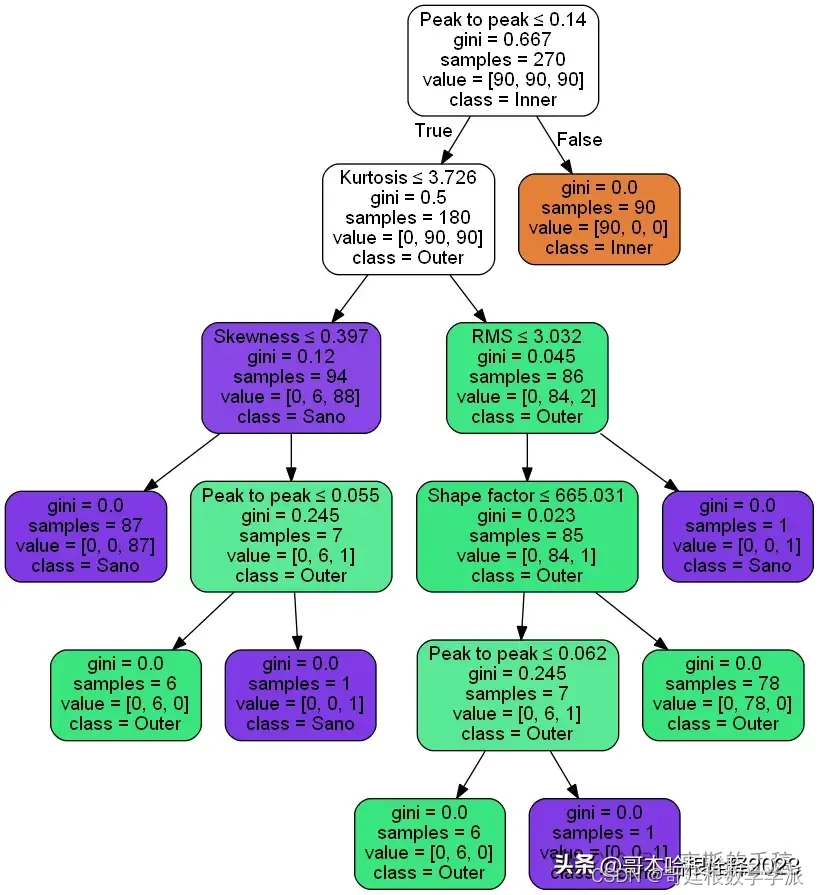
from sklearn.tree import export_graphviz
from six import StringIO
from IPython.display import Image
import pydotplus
dot_data = StringIO()
estadisticos = ['Kurtosis', 'Impulse factor', 'RMS', 'Margin factor', 'Skewness', 'Shape factor', 'Peak to peak', 'Crest factor']
export_graphviz(treeModel, out_file=dot_data, filled=True, rounded = True,special_characters = True, feature_names = estadisticos, class_names = ['Inner', 'Outer', 'Sano'])
graph = pydotplus.graph_from_dot_data(dot_data.getvalue())
graph.write_png('tree_hamming.png')
Image(graph.create_png())