Title
题目
Cascade multiscale residual attention CNNs with adaptive ROI for automatic brain tumor segmentation
自适应感兴趣区域的级联多尺度残差注意力CNN用于自动脑肿瘤分割
01
文献速递介绍
脑肿瘤是大脑细胞异常和不受控制的增长,被认为是神经系统中最具威胁性的疾病之一。根据美国国家脑肿瘤协会(NBTS)的数据,全球每年约有40万人患上脑肿瘤,每年约有12万人死亡,而且这一数字还在逐年增加。成人最常见的原发性脑肿瘤是胶质瘤,会对中枢神经系统造成严重损害。胶质瘤始于大脑的胶质细胞,通常分为低级别胶质瘤(LGG)和高级别胶质瘤(HGG)。HGG比LGG更为恶性,因其增长迅速,患者的平均预期寿命约为两年。
随着医学影像技术的发展,例如磁共振成像(MRI),脑肿瘤可以通过磁共振的各种序列成像,例如T1加权、T2加权、带对比增强的T1(T1c)和液体衰减反转恢复(FLAIR)图像来成像,如图1所示。准确的脑肿瘤分割对于医学诊断、治疗规划和手术规划至关重要。然而,由于形状、大小以及脑肿瘤定位的巨大变异,脑肿瘤的自动分割是一项具有挑战性的任务。具体而言,核心和增强肿瘤的分割更具挑战性,因为相邻结构之间的边界不清晰,并且具有平滑的强度梯度。
在过去的几十年里,已经进行了大量关于自动脑肿瘤分割的研究。然而,基于深度学习(DL)的技术的最新进展已经显示出对各种与计算机视觉相关的问题以及特别是与健康相关的问题的性能显著提高。DL技术已被广泛应用于脑肿瘤分割任务,以提高整个肿瘤(WT)、增强肿瘤(ET)和肿瘤核心(TC)区域的性能。这些技术已经表现出对整个肿瘤组织的分割性能。不幸的是,由于其较小的尺寸和与整个肿瘤相似的纹理,对增强和核心肿瘤组织的准确分割仍然是一个具有挑战性的问题。因此,所有现有的技术均无法像对整个肿瘤区域那样为核心和增强肿瘤组织提供相同的性能。
在基于DL的技术中,3D卷积神经网络(CNNs)被认为更适合于体积分割任务,这比2D CNNs计算上更昂贵。而2D-CNN以2D方式探索每个切片中的肿瘤存在,并且需要较少的计算资源和训练样本。然而,2D-CNN无法处理3D顺序信息,这对于体积分割至关重要,因此会影响分割性能。为了兼顾两种体系结构,我们采用了一种折衷方法,作为混合方法,可以在利用三维CNNs的多少内存的情况下利用间隔切片的顺序信息。
我们提出了一种基于自适应感兴趣区域(A-ROI)的级联多尺度残差注意力UNet(MRA-UNet)架构,用于分割脑肿瘤区域(即整个、增强和核心肿瘤)。所提出的方案通过使用A-ROI策略[8]显着提高了增强和核心肿瘤的分割性能,使其能够仅专注于整个肿瘤区域而丢弃多余的信息。与传统的3D CNNs相比,MRA-UNet在计算上更高效,因为它仅以三个连续切片作为输入来分割肿瘤。我们还研究了诸如条件随机场(CRF)和测试时间增强(TTA)之类的后处理策略以改善分割性能。所提出的工作已在BraTS2017、BraTS2019和BraTS2020数据集上进行了严格评估。与现有的最新技术相反,所提出的技术在整个脑肿瘤的分割上取得了有竞争力的性能,同时在增强和核心肿瘤分割方面表现出了显著的优势。
Abstract
摘要
A brain tumor is one of the fatal cancer types which causes abnormal growth of brain cells.Earlier diagnosis of a brain tumor can play a vital role in its treatment; however, manualsegmentation of the brain tumors from MRI images is a laborious and time-consumingtask. Therefore, several automatic segmentation techniques have been proposed, but highinter-and intra-tumor variations in shape and texture make accurate segmentation ofenhanced and core tumor regions challenging. The demand for a highly accurate and robustsegmentation method persists; therefore, in this paper, we propose a novel fully automatictechnique for brain tumor regions segmentation by using multiscale residual attentionUNet (MRA-UNet). MRA-UNet uses three consecutive slices as input to preserve thesequential information and employs multiscale learning in a cascade fashion, enabling itto exploit the adaptive region of interest scheme to segment enhanced and core tumorregions accurately. The study also investigates the application of postprocessing techniques (i.e., conditional random field and test time augmentation), which further helpsto improve the overall performance. The proposed method has been rigorously evaluatedon the most extensive publicly available datasets, i.e., BraTS2017, BraTS2019, andBraTS2020. Our approach achieved state-of-the-art results on the BraTS2020 dataset withthe average dice score of 90.18%, 87.22%, and 86.74% for the whole tumor, tumor core, andenhanced tumor regions, respectively. A significant improvement has been demonstratedfor the core and enhancing tumor region segmentation showing the proposed method’sevident effectiveness.
脑肿瘤是一种致命的癌症类型,会导致大脑细胞异常增长。早期诊断对脑肿瘤的治疗起着至关重要的作用;然而,从MRI图像中手动分割脑肿瘤是一项费时费力的任务。因此,提出了几种自动分割技术,但是形状和纹理的高度差异使得增强和核心肿瘤区域的准确分割具有挑战性。对高度准确和稳健的分割方法的需求依然存在;因此,在本文中,我们提出了一种新颖的全自动脑肿瘤区域分割技术,使用多尺度残差注意力UNet(MRA-UNet)。MRA-UNet使用三个连续的切片作为输入以保留顺序信息,并采用级联多尺度学习,使其能够利用自适应感兴趣区域方案准确地分割增强和核心肿瘤区域。该研究还调查了后处理技术的应用(例如,条件随机场和测试时间增强),进一步帮助提高了整体性能。所提出的方法在最广泛公开的数据集上进行了严格评估,即BraTS2017、BraTS2019和BraTS2020。我们的方法在BraTS2020数据集上取得了最先进的结果,对于整个肿瘤、肿瘤核心和增强肿瘤区域的平均Dice分数分别为90.18%、87.22%和86.74%。对于核心和增强肿瘤区域分割显示出了所提方法的明显有效性,取得了显著的改进。
Method
方法
The proposed method follows the typical automatic brain tumor segmentation workflow as demonstrated in Fig. 2. However, at each stage of workflow, significant changes have been introduced to improve the final segmentation performance.First, pre-processing is performed to produce focused and enhanced images, having improved contrast for brain tumorregions. Also, to preserve the sequential information, the three consecutive slices have been concatenated to create threechannel images like typical RGB images. Secondly, brain tumor segmentation is performed by using MRA-UNet architecturein a cascade fashion, which provides segmentation of whole, core, and enhanced tumor regions. We apply the novel A-ROIapproach [8] which helps cascade architecture to focus on core and enhanced tumors. Finally, we perform postprocessing,which further refines the output of MRA-UNet by using postprocessing techniques (i.e., CRF and TTA). The detail of each stepis provided in the following subsections.
所提出的方法遵循了典型的自动脑肿瘤分割工作流程,如图2所示。然而,在工作流程的每个阶段,都引入了显著的变化以提高最终的分割性能。
首先,进行预处理以生成焦点和增强图像,对脑肿瘤区域的对比度进行改善。此外,为了保留顺序信息,将三个连续的切片连接起来,创建了三通道图像,类似于典型的RGB图像。其次,采用级联式MRA-UNet架构进行脑肿瘤分割,提供整个、核心和增强肿瘤区域的分割。我们采用了新颖的A-ROI方法[8],有助于级联架构专注于核心和增强的肿瘤。最后,通过使用后处理技术(即CRF和TTA),对MRA-UNet的输出进行进一步的精化。每个步骤的详细信息将在接下来的子节中提供。
Conclusion
结论
This study proposes a novel technique for brain tumor segmentation and its classification into a whole, enhanced, andcore tumor region. The proposed method employs multiscale residual attention convolutional neural networks (MRA UNet) architecture in a cascade fashion with the adaptive region of interest (A-ROI). The technique takes the three consecutive slices as input to incorporate the sequential information, which significantly improves the whole tumor segmentationaccuracy, and the application of adaptive-ROI significantly improves the segmentation performance for enhanced and coretumor segmentation. Finally, the work investigates the effect of postprocessing strategies such as conditional random fieldand test time augmentation to improve the segmentation. The proposed work has been rigorously evaluated on theBraTS2017, BraTS2019, and BraTS2020 datasets. The comparison with the existing state-of-the-art methods shows thatthe proposed techniques achieved competitive performance for whole-brain tumors and significantly outperformed forenhanced and core tumor segmentation. Future plans include the extension of MRA-UNet with deep supervised learningto enable the end-to-end learning to avoid the cascade mechanism without degrading the performance for enhanced andcore tumor regions.
本研究提出了一种新颖的技术,用于脑肿瘤的分割和其分类为整个、增强和核心肿瘤区域。所提出的方法采用了级联式的多尺度残差注意力卷积神经网络(MRA UNet)架构,结合自适应感兴趣区域(A-ROI)。该技术将三个连续的切片作为输入,以融合顺序信息,从而显著提高了整个肿瘤分割的准确性,并且自适应感兴趣区域的应用显著提高了增强和核心肿瘤分割的性能。最后,本研究调查了条件随机场和测试时间增强等后处理策略对分割结果的影响。所提出的方法在BraTS2017、BraTS2019和BraTS2020数据集上进行了严格评估。与现有最先进方法的比较显示,所提出的技术在整个脑肿瘤方面取得了具有竞争力的性能,并且在增强和核心肿瘤分割方面表现出显著优势。未来计划包括将MRA-UNet扩展到深度监督学习,实现端到端学习,避免级联机制而不降低增强和核心肿瘤区域的性能。
Results
结果
In this section, firstly, we have analysed the overall performance, secondly, we have compared the performance of ourproposed methodology with the state-of-the-art methods of brain tumor segmentation approaches on BraTS 2020, BraTS2019, and BraTS 2017 datasets. The performance has been validated quantitatively, as well as visual analysis has been performed and compared with existing state-of-the-art techniques. Thirdly, we have carried out the ablation studies to showthe validity of each contribution presented in this work. The detailed analysis is provided in the following subsections.
在本节中,首先,我们对整体性能进行了分析,其次,我们将我们提出的方法与BraTS 2020、BraTS 2019和BraTS 2017数据集上的脑肿瘤分割方法的最新技术进行了比较。性能已经通过定量验证,并进行了可视化分析,与现有的最新技术进行了比较。第三,我们进行了消融研究,以展示本工作中提出的每项贡献的有效性。详细分析见以下子节。
Figure
图
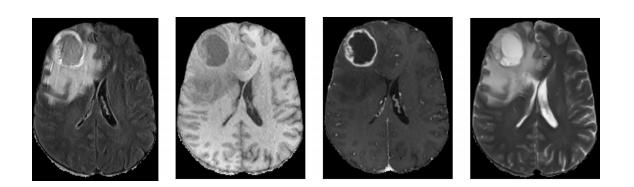
Fig. 1. Brain MRI data from BraTS 2020 database. From left to right: FLAIR, T1, T1ce, and T2 slices.
图1. 来自BraTS 2020数据库的脑MRI数据。从左到右:FLAIR、T1、T1ce和T2切片。
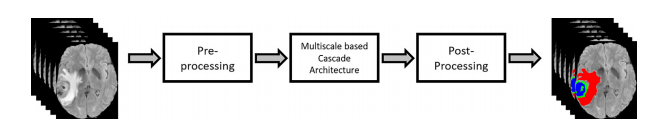
Fig. 2. Overview of the proposed method.
图2. 所提出方法的概述。
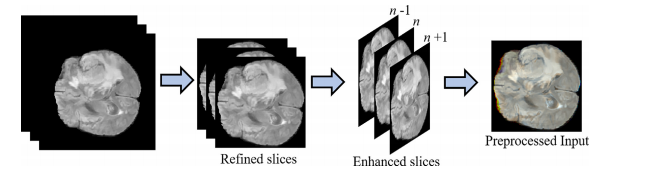
Fig. 3. Preprocessing stage including scan refinement, image enhancement and slice concatenation.
图3. 预处理阶段,包括扫描细化、图像增强和切片串联。
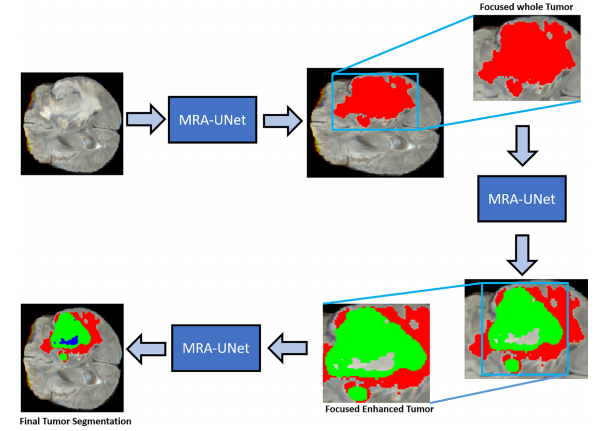
Fig. 4. Illustration of our proposed multiscale residual attention-based hourglass-like architecture
图4. 我们提出的基于多尺度残差注意力的沙漏式结构示意图
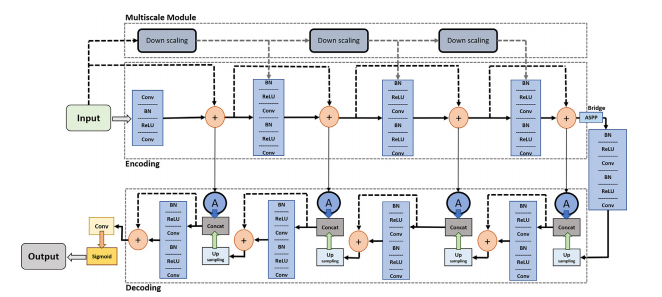
Fig. 5. Block diagram of the proposed MRA-UNet architecture.
图5. 所提出的MRA-UNet架构的块图。
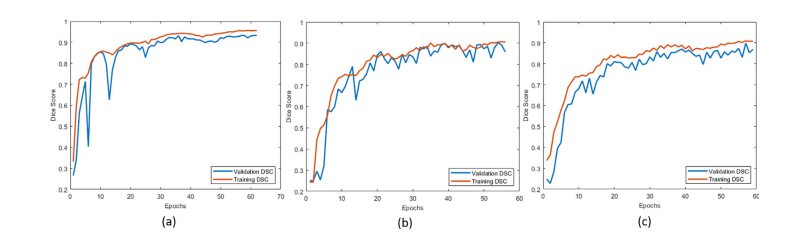
Fig. 6. Learning curves are shown in (a), (b) and (c) for whole, core and enhanced tumor segmentation, respectively
图6. 学习曲线如下所示:(a) 整个肿瘤分割,(b) 肿瘤核心分割,(c) 强化肿瘤分割。
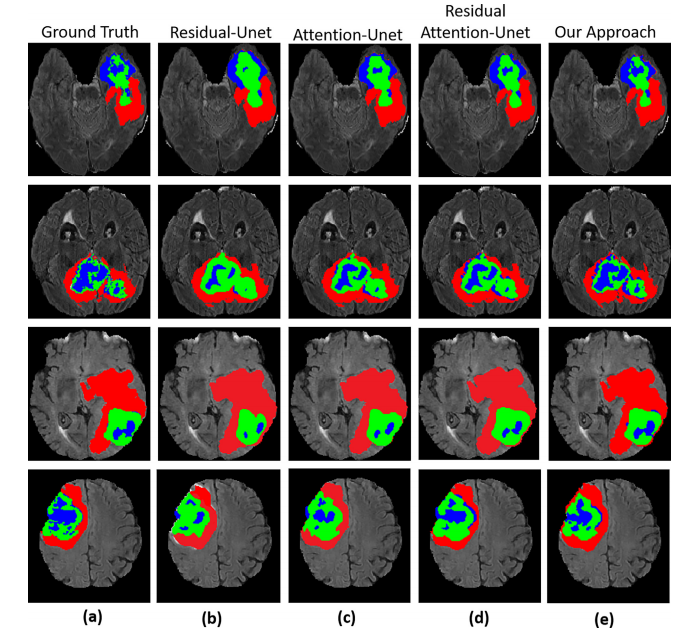
Fig. 7. Segmentation results of the whole tumor, enhancing tumor, and tumor core. (a) represents Ground truth (label), (b) Residual-UNet (c) AttentionUNet (d) Residual Attention-UNet (e) Our method.
图7. 整个肿瘤、增强肿瘤和肿瘤核心的分割结果。(a) 表示地面真相(标签),(b) 残差UNet,(c) 注意UNet,(d) 残差注意力UNet,(e) 我们的方法。
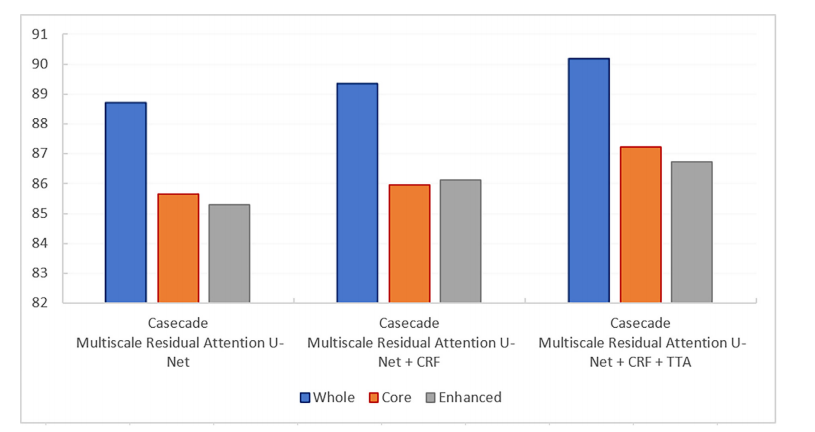
Fig. 8. Comparison of proposed cascade MRA-UNet strategy without and with post processing techniques.
图8. 比较了未经后处理技术和经过后处理技术的提出的级联MRA-UNet策略。
Table
表

Table 1Total number of scans for training, testing, and validation of the three datasets.
表1三个数据集的训练、测试和验证扫描总数。
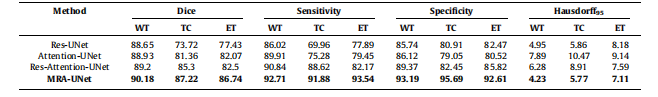
Table 2Mean standard deviation for quantitative results of different conventional networks with proposed methodology on the BraTS2020 dataset, the bestperformance indicated in bold. WT - whole tumor, TC - tumor core, ET - enhancing tumor.
表2:使用所提出的方法在BraTS2020数据集上对不同传统网络进行定量结果的均值和标准差,最佳性能用粗体表示。WT - 整个肿瘤,TC - 肿瘤核心,ET - 强化肿瘤。
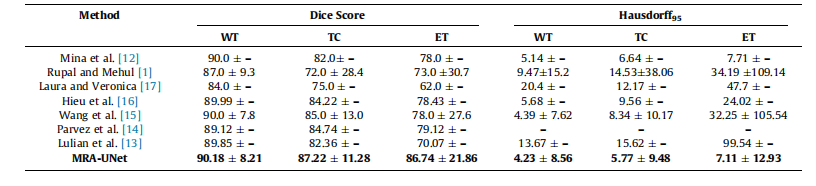
Table 3Mean standard deviation for quantitative results of various segmentation methods on the BraTS2020 dataset, the best performance indicated in bold. WT -whole tumor, TC - tumor core, ET - enhancing tumor.
表3:BraTS2020数据集上各种分割方法的定量结果的均值和标准差,最佳性能用粗体表示。WT - 整个肿瘤,TC - 肿瘤核心,ET - 强化肿瘤。
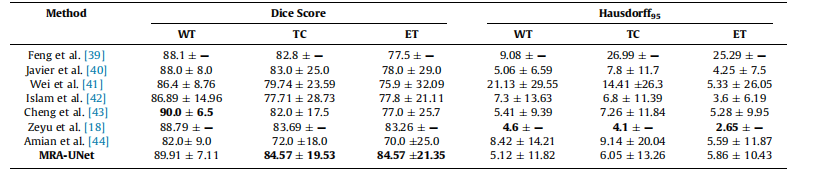
Table 4Mean standard deviation for quantitative results of various segmentation methods on the BraTS2019 dataset, the best performance indicated in bold.
表4:BraTS2019数据集上各种分割方法的定量结果的均值和标准差,最佳性能用粗体表示。
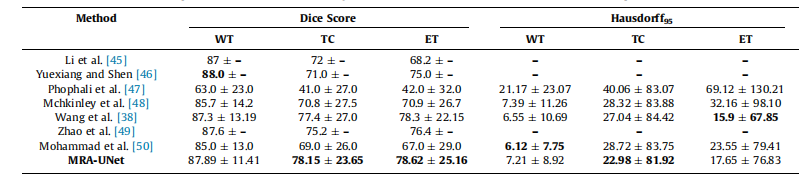
Table 5Mean standard deviation for quantitative results of various segmentation methods on the BraTS2017 dataset, the best performance indicated in bold.
表5:BraTS2017数据集上各种分割方法的定量结果的均值和标准差,最佳性能用粗体表示。
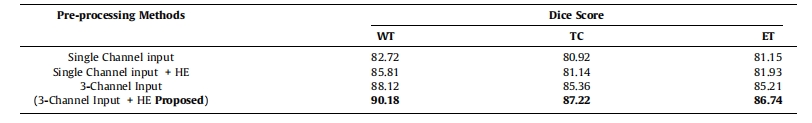
Table 6Pre-processing using different channels with, and without histogram equalization.
表6:使用不同通道进行预处理,包括直方图均衡化和不均衡化的效果对比。


)




